Quick Links
General Questions
What is MiDOG®?
MiDOG® is a state-of-the-art testing facility employing the latest, cutting-edge, molecular-based and Next-Generation DNA Sequencing technologies to accurately identify and characterize microbial populations in collected samples. The presence of both bacterial and fungal pathogens as well as antibiotic susceptibility (resistance) are revealed using the proprietary pipeline developed for the MiDOG® service offering.
Want to learn more? Watch our summary video here!
What is the MiDOG® All-in-One Microbial Test?
The test is comprised of two parts:
1) A swab collection kit for sampling done by the veterinarian and 2) a Next Generation DNA sequencing service performed at the MiDOG® testing facility on the samples submitted by the veterinarian.
What does the MiDOG®All-in-One Microbial test provide to the veterinarian?
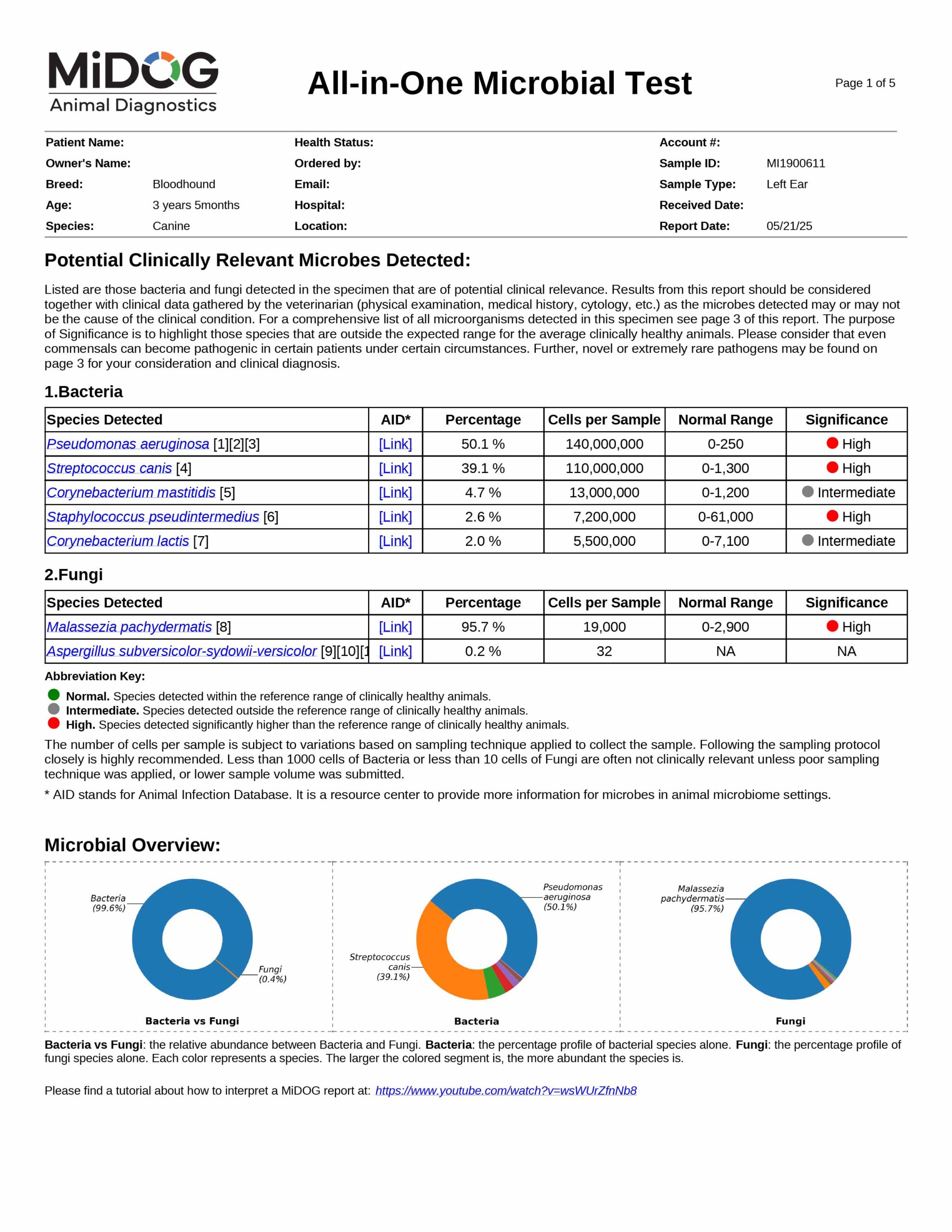
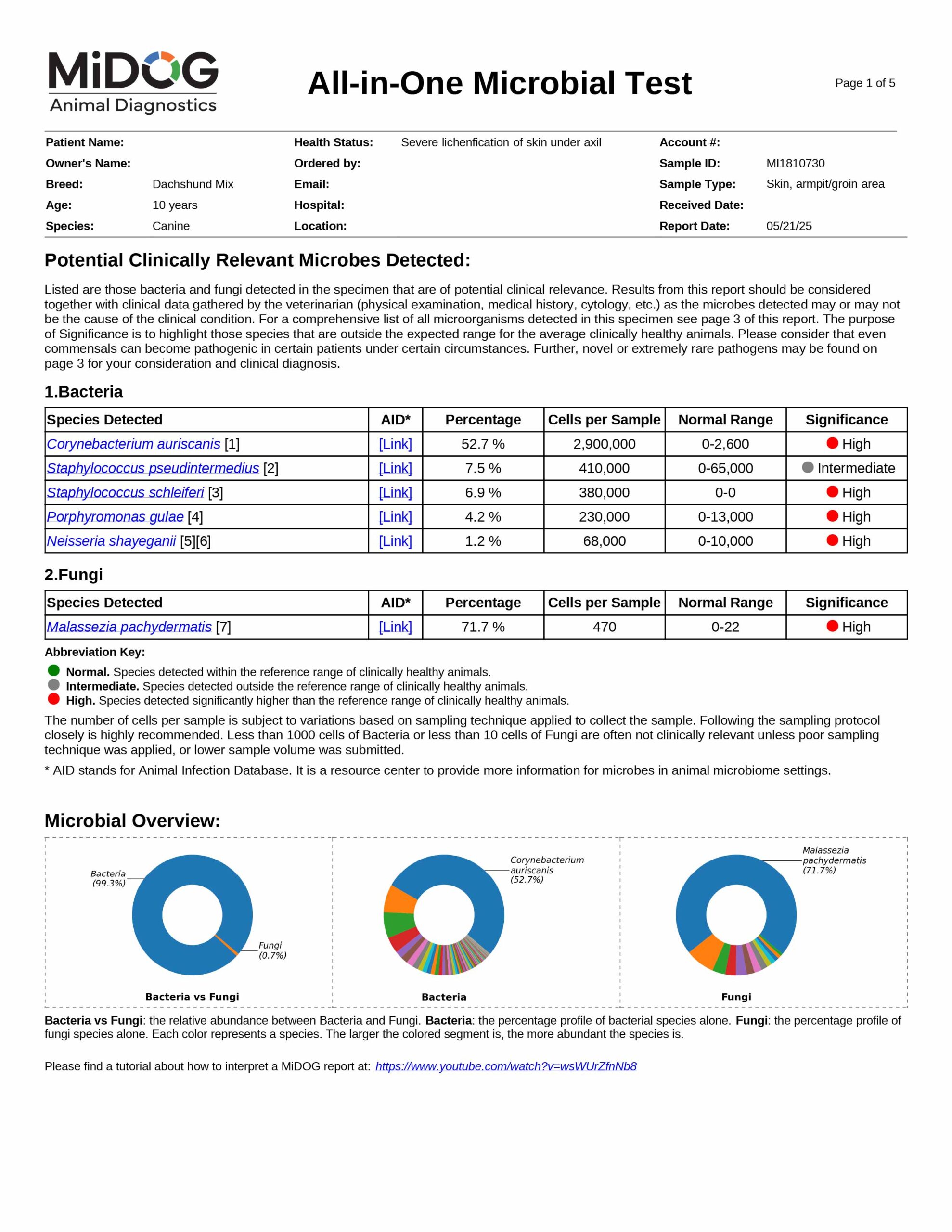
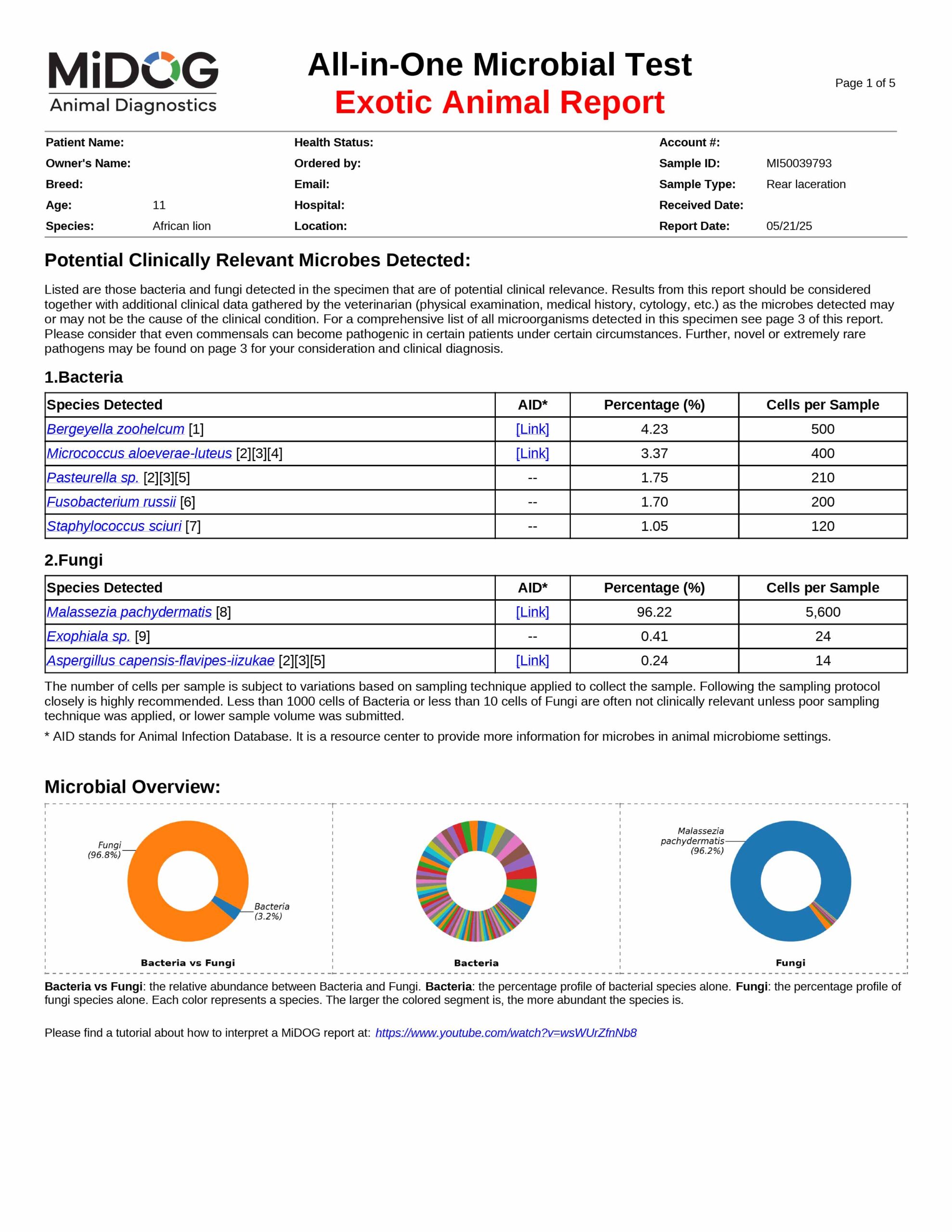
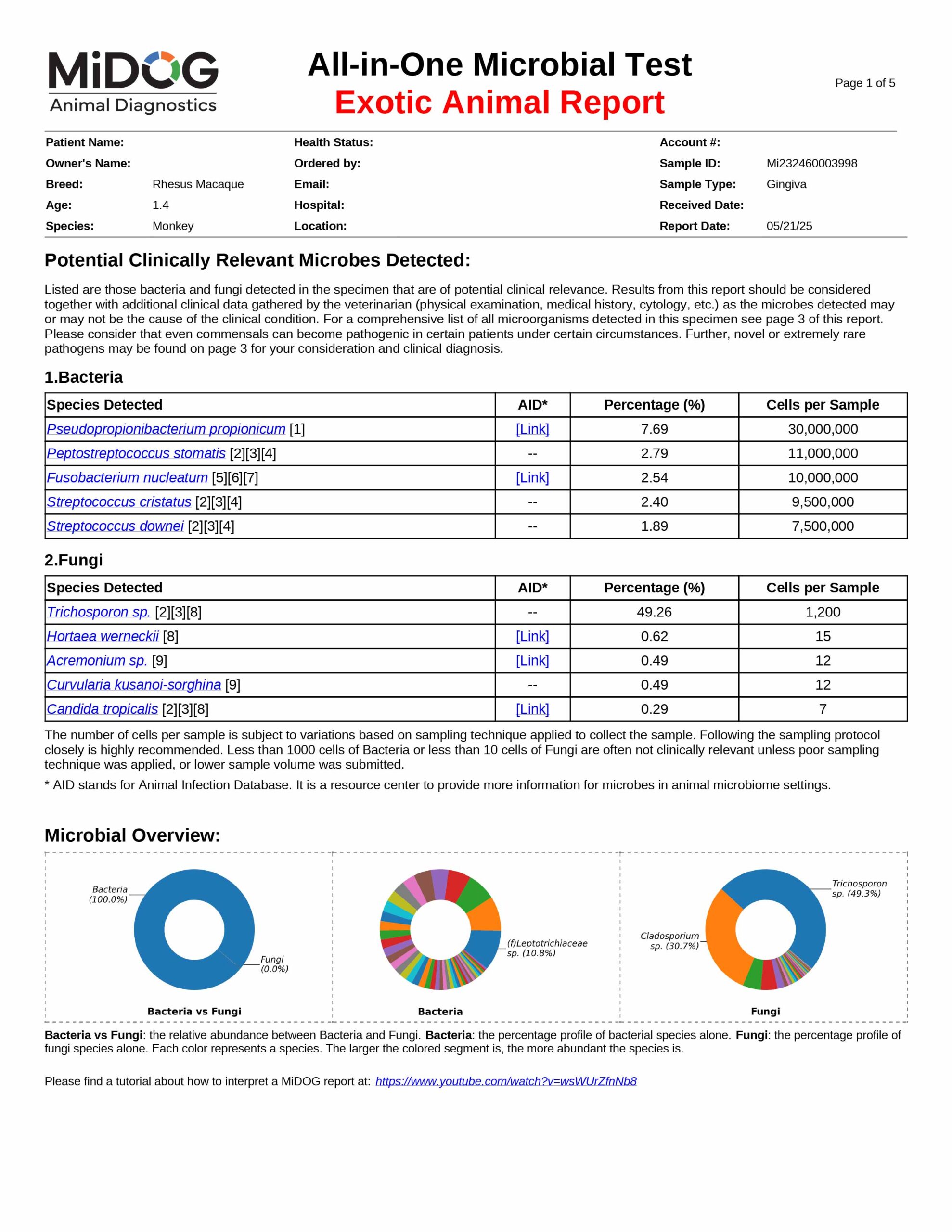
The veterinarian is provided a comprehensive report outlining the results of the test. The report consists of three parts:
- Bacterial and fungal pathogens detected.
- Antibiotic resistance profile for each identified bacterial pathogen.
- Supplemental data: Anything unusual that we find when comparing the sample to our clinically healthy canine database, as well as supplemental data on bacterial and fungal profile
All in just one test! A sample report is shown here.
Watch a video explaining the different components of the report here.
What are the benefits of using this test?
99% of all organisms cannot currently be cultured in the lab and are thus missed upon analysis. Compared to culturing tests, this test tells you much faster about ALL of the organisms that are present on your sample, and how your patient might respond to treatment. This can significantly shorten the time that your patient is suffering from infections or lesions and aid effective antibiotic stewardship.
Where can I find a list of the organisms covered by this test?
There is currently no definitive list available because the MiDOG All-in-One Test detects up to 123,701 organisms, including fungi, bacteria, protozoa, and parasites. This test uses next-generation DNA sequencing, a highly sensitive, molecular-based diagnostic tool.
The All-in-One Test targets two highly conserved genomic regions: the 16S rRNA region for bacteria and the ITS region for fungi. Because every bacterial and fungal species contains these regions, the test can detect virtually all bacterial and fungal species present in a sample. This advanced sequencing method identifies and quantifies each organism and determines their antibiotic resistances.
After sequencing, the DNA data is compared against extensive genomic databases to identify species and assess whether they are known pathogens. Current databases contain billions of sequences covering more than 450,000 documented species. If a sequence is not yet described, the test identifies the closest known relative.
In short, this means the MiDOG All-in-One Test can detect virtually any bacterial or fungal species of interest present in the sample.
Can I submit a sample from a patient that is currently on antibiotic treatment?
Yes, we can test patients that are currently taking antibiotics. We are able to see if the pathogen(s) are responding to the treatment, if there may be a secondary pathogen that is resistant to the antibiotic, and/or if commensals are impacted by the treatment as well. Especially for skin or ear swabs or feces, this is a great approach to track the real time response of the microbiome and pathogens to the antibiotic.
Ideally, the veterinarian would collect a sample before the treatment to select the most appropriate antibiotic (if needed), during the antibiotic treatment to see if the pathogen is responding to the treatment, and afterwards to see if the infection has cleared. Commensals may also be impacted by the treatment, and a lack of microbial diversity after an antibiotic treatment may be suggesting the use of probiotics in some cases. This approach has been previously shown in the Journal of Investigative Dermatology, Bradley et al. 2016 (https://www.sciencedirect.com/science/article/pii/S0022202X16004553)
How long after an antibiotic treatment should I wait to submit a sample?
There is no one-fits-all answer for this question. It depends on the sample type, the antibiotic treatment, the dose, and the question the clinician would like to answer. Please also refer to question “Can I submit a sample from a patient that is currently on antibiotic treatment?”. The amount of time an antibiotic can stay in the system can vary greatly (between hours and days), and the time for the commensal/healthy microbiome population to go back to ‘normal’ varies accordingly. A rule of thumb is, if the patient is no longer showing clinical signs of infection, please wait one week to submit a check up sample.
Sample Collection and Report Questions
Accepted Sample Types
All samples must be submitted using the official MiDOG collection tubes, no exceptions. Urine samples must be collected in the MiDOG urine collection tube (yellow label), while all other sample types must be placed in the MiDOG collection tubes with white labels.
Any samples not submitted in MiDOG collection tubes will be rejected and discarded. Please do not include any additional materials outside the designated collection tubes.
MiDOG accepts all types of animal samples, including blood. For blood samples, we accept both serum and plasma, with an ideal volume of 0.5–1 ml.
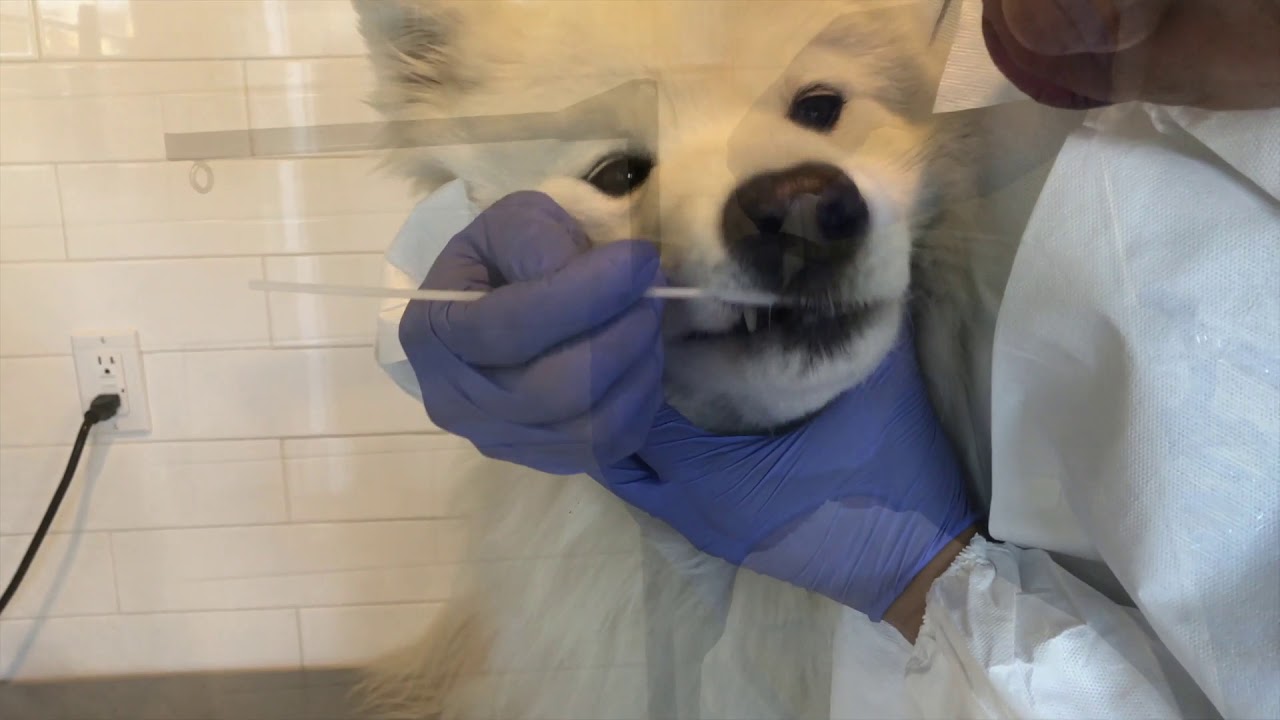
How do I collect a sample for the MiDOG® Service?
Follow the instructions provided in the MiDOG® All-in-One Microbial Test manual. The Swab Collection Manual can also be found here, and the Urine Collection Manual can be found here or watch our MiDOG® sample collection instruction video here. Please don’t hesitate to contact us should you have any questions.
How do I get my MiDOG® results?
The results will be sent to the veterinarian via email in PDF format. A sample report is shown on the right.
Where should I send my samples?
We provide you with a prepaid and pre-labelled FedEx envelope to send your samples to us. Our laboratory address is
MiDOG Laboratory, 14762 Bentley Cir., Tustin CA 92780.
Do I need to ship the samples on ice?
No, there is no need to store or ship your samples on ice.
How long will it take until I get the report?
Our turn around time is two to five business days from when we receive the sample. We will email the report to the veterinarian.
What is the shelf life of the MiDOG® Microbial Test kit?
12 months.
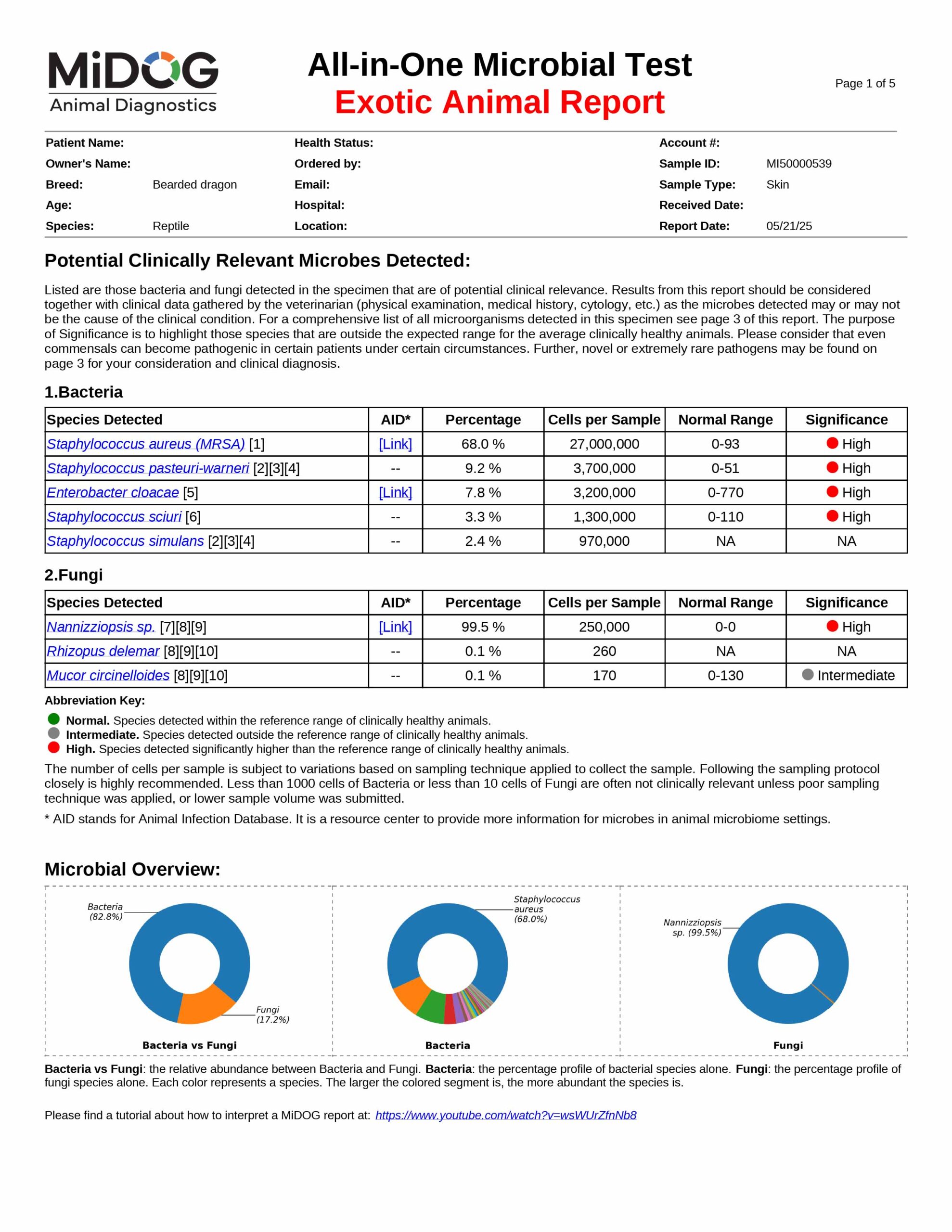
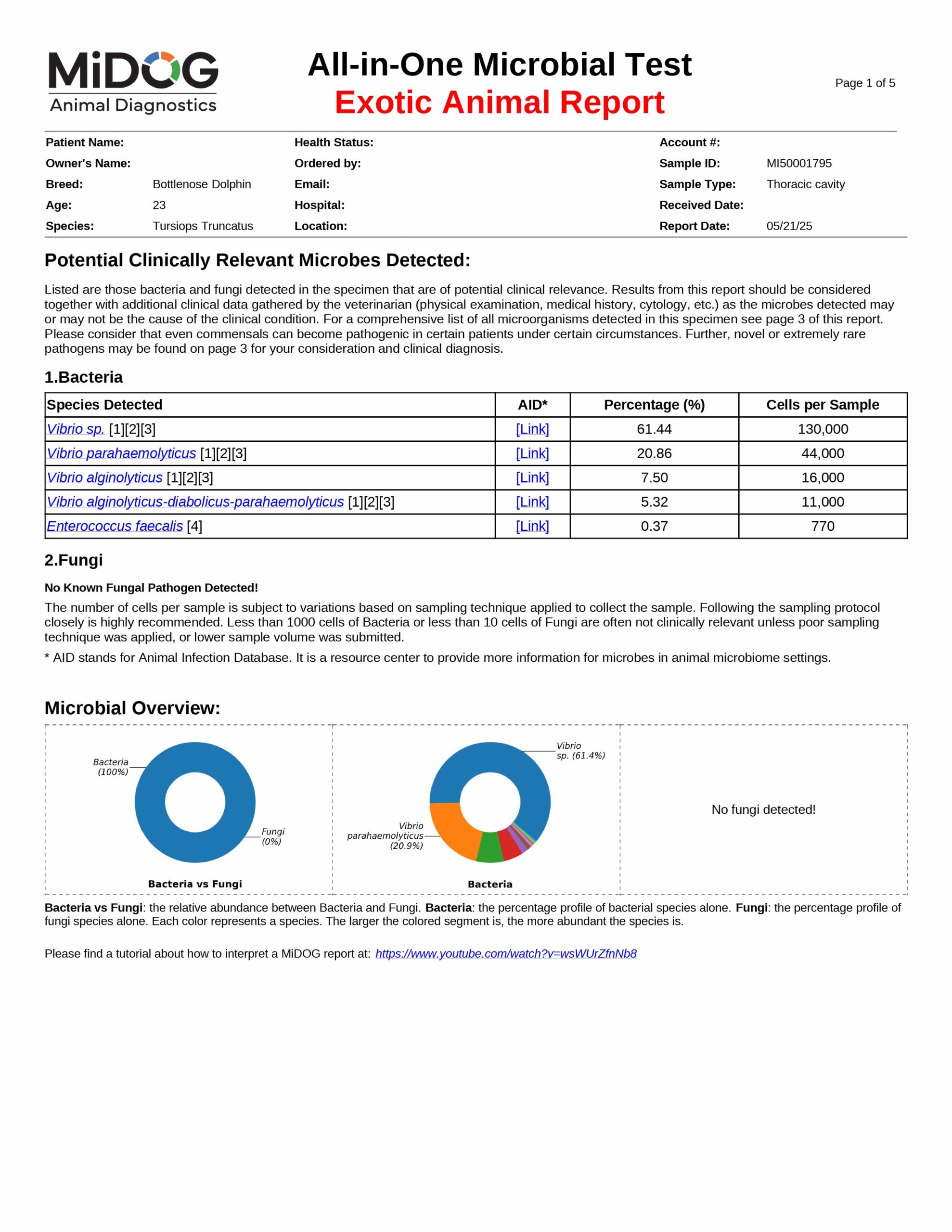
Can I send samples from other species besides canines as well?
Yes, absolutely. The only difference is that we cannot compare your sample to a clinically healthy database.
I sampled my patient and forgot to send the sample in. Is the sample still OK?
Yes, it is. The DNA preservation reagent we provide you with stabilizes your sample at room temperature for up to one year.
Can I send multiple samples in one envelope?
Yes, please send multiple samples in one envelope.
Can I submit fluid samples?
Yes, MiDOG can test a wide variety of fluid samples, including blood. We accept both serum and plasma, with an ideal volume of 0.5–1 ml. Other acceptable fluid types include bodily fluids (e.g., bile, joint fluid, urine), washes (e.g., eye washes, nasal washes), or rinses. We require a minimum of 2 ml of liquid for testing, but 4 ml is recommended to ensure optimal accuracy.
Please ensure that the fluid sample is placed directly into the collection tube; do not submit swabs soaked in liquid.
For fluids, use MiDOG collection tubes with the white label. For urine samples, please use the dedicated urine collection kits, which have yellow-labeled tubes.
For more information, please call customer service at (833) 456-4364.
Can I submit less than 2ml of fluid samples?
If only less than 2 ml of fluid can be collected from a patient, it is still possible to submit the sample. While a larger sample volume would be ideal to ensure accuracy of the analysis, we will process even samples with smaller volumes.
Please note, the detection limit of the MiDOG All-in-One assay is 200 cells per sample. If less than 2ml of submitted, the report may not list any bacteria or fungi.
Can I submit water or pool samples?
Yes, it is possible to test pool or aquarium water samples as well. Ideally, 4ml of water sample is mixed with the MiDOG collection tubes with the white label. Simply close the collection tube, invert the sample tube 5 to 10 times, and send it to the MiDOG laboratory with the provided return envelopes. It is possible to use the provided sterile swab to take a swab sample of the pool filter as well. You may add the swab to the sample same collection tube as the pool water. If two separate samples are submitted, there would be a charge for two samples.
Can I submit tissue samples?
Yes, you may submit animal tissue samples. To ensure the preservation of the material, please grind up the tissue in a sterile mortar and pestle before adding it to the MiDOG collection tube with the white label.
Please do not send any additional or extra material. All sampling material must be inside the MiDOG collection tube. Any samples that are not provided using the MiDOG collection tubes will be discarded.
Can I submit samples of body parts, for example a toe, a leg, or tail?
Only samples inside the MiDOG collection tubes will be accepted. To ensure the preservation of the material, please grind up the sample in a sterile mortar and pestle before adding it to the MiDOG collection tube with the white label.
For urine samples, is cystocentesis or free catch preferred? Will contaminant bacteria show on free catch?
Both collection methods are accepted. However, a sample collected via cysto is preferred. A cysto sample will only contain bacteria and fungi found in the urine, while a free catch urine sample can also contain bacteria and fungi originated from the skin as the urine is being released. Cysto samples will only show pathogens in the urine and will not contain potential skin contaminants
Our reports highlight all pathogens on the first page, so members of the normal skin microbiome would only appear in the supplemental information on the third page. Further, we include the cell numbers for each pathogen and commensal organism detected. The cell numbers will also aid in differentiating between a clinically relevant organisms and a commensal coming from the skin. You can view our example report walkthrough here to see what’s included on each page of the report. If you have any questions on how to interpret your report, we are happy to offer you free veterinary consultation to help with treatment decisions.
What is the difference between a small-sized swab and a regular one?

Ordering
Where can I order the MiDOG® Canine Microbial Test?
To order a sample kit simply go to the ‘ORDER’ tab on this website or send us an email to orders@midogtest.com.
Please note that the MiDOG® Canine Microbial Test is only provided to veterinarians.
How many swabs are included in MiDOG® All-in-One Microbial Test Swab Collection Kit?
Five swabs and five collection tubes are included.
Do I have to pay for shipping?
The collection kits are free of charge, only a shipping fee of $17.50 is applied.
Science
What is Next Generation Sequencing?
Next Generation Sequencing (NGS) is a molecular based method that can sequence and detect the DNA of ALL microbes that live on your dog’s skin, without having to grow any of these organisms in a laboratory first. NGS is considered the gold standard for the detection and identification of microorganisms.
More information about NGS can be found on Illumina’s® webpage.
Portal Questions
Where can I find my login information?
Your login information was provided to you via email from our webservices department. If you cannot locate this information or need to reset your password, please send us an email to info@midogtest.com.
Where can I find my account number?
Click the MiDOG tab on the top right-hand corner above your clinic name.
How can I add a user to my existing account?
- Click the MiDOG tab on the top right-hand corner above your clinic name.
- Click “Create new user” on the left side of the screen.
- Once all the information is completed Click “Create user” to save the information.
Do I have to set up a new account if working from multiple clinics?
Yes, every clinic or hospital has their own account number, login information, and billing information.
Where can I update my clinic information, like mail address, shipping address, billing information, etc.?
Click the MiDOG tab on the top right-hand corner above your clinic name, then click the green tab button “Edit Account Setting” to change or add the information.
I forgot the account password, what do I do now?
Contact the MiDOG team by email or phone at info@midogtest.com or (833) 456-4364.
Can I send samples from other species besides canines as well?
Yes, absolutely. The only difference is that we cannot compare your sample to a clinically healthy database.
I sampled my patient and forgot to send the sample in. Is the sample still OK?
Yes, it is. The DNA preservation reagent we provide you with stabilizes your sample at room temperature for up to one year.
How do I place an order?
Go to the “Supplies” tab on the left top side. Once on the order screen, please select the desired product and “add to cart” to complete your new order.
Is there a limit on how many test kits can be ordered?
You can order up to 5 test kits (a total of 25 test). If you need more supplies, please contact customer service via email info@midogtest.com or over the phone (833) 456-4364.
What types of collection kits are available?
Swab, Small-sized Swab, and Urine collection kits.
How many test preps are in each kit?
There are 5 test preps in each kit.
Where can I find pricing information?
Pricing information can be requested via email info@midogtest.com or over the phone (833) 456-4364.
How can I track an order?
Your order can be tracked by the tracking number provided via email or you can call the MiDOG Customer Service department at (833) 456-4364.
How do I order more return envelopes?
You can email info@midogtest.com or call (833) 456-4364.
Where do I pay my bill?
You can pay your bill online through the MiDOG portal, or by following the link below, by clicking the link in the email that we sent with your invoice information, over the phone at (833) 456-4364, or by mailing a check to MiDOG LLC, 14762 Bentley Cir., Tustin CA 92780.
How can I update my billing information?
Please log into your account and update your billing information under the “PROFILE” option on the far-right hand side. Otherwise, please send us an email to info@midogtest.com or give us a call at (833) 456-4364.
Can I sign up for automated payments?
Yes, you can put your credit card on file for automated payments.
How do I collect my patient’s sample?
Instructions on how to collect samples are included inside of every kit, can be found on our website in writing or video. After you have collected the sample using the MiDOG collection kit, simply send it to our laboratory for testing.
What type of samples can MiDOG test?
MiDOG can test any sample types, except blood.
I submitted a sample, but it shows as not received. Where can I get more information?
You can track it the FEDEX return labels provided with your order. We recommend always noting this information before sending us a sample for faster tracking. If you have not recorded the tracking number used, you can call our customer service department and they can help you locate the tracking number.
How do I enter patients’ information?
Once you have placed an order the system will automatically give you the tube identification numbers sent with the order. When you are ready to collect a sample, log onto your account, and click the “Labs” tab on the top left corner. You can then enter the information corresponding to the tube you will be using. Sample tubes that have not been used are marked as “MISSING TUBE ID”. Once all information has been added make sure to click “save changes”.
How do I locate my patient’s report and view my report?
To view your patient’s report, click “LABS” on the top left-hand side of the screen. Enter either the “Tube Number, “Patient Name”, or owner name to filter the search results.
How do I know if a sample was received?
You will receive an email with the patient information letting you know that we have received your sample. You can also view status by clicking the “LABS” tab. Information is updated every business day.
How do I check the status of a sample?
You can view the status by clicking the “LABS” tab.
How do I view older results?
To view your patient’s past results, click “LABS” on the top left-hand side of the screen. Enter either the “Tube Number, “Patient Name”, or owner name to filter the search results.
How do I download my results?
To download your patient’s past results, click “LABS” on the top left-hand side of the screen.
Where can I find my results?
To view your patient’s report, click “LABS” on the top left-hand side of the screen. Enter either the “Tube Number, “Patient Name”, or owner name to filter the search results.
Can I search my reports for a certain patient?
Yes. To search your patient’s report, click “LABS” on the top left-hand side of the screen.
Enter either the “Tube Number, “Patient Name”, or owner names to filter the search results.
How do I interpret the results?
You may find an example video here. If you have additional questions, please give us a call at (833) 456-4364.
How do I schedule a consultation?
You may schedule a consultation using the portal by clicking on the consultation tab. Otherwise, please send us an email or give the customer service department a call at (833) 456-4364.
Can’t find what you are looking for?
Please contact us at info@midogtest.com or give us a call at (833) 456-4364. If you still need furtehr portal instructions, please view the Portal Navigation Video here.